Amplicon sequencing: a quick look at the construct
In short, amplicon sequencing refers to the technology that amplifies (makes several copies of) a target (genomic) region and then sequences it. A common example is metataxonomics (Marchesi & Ravel 2015: The vocabulary of microbiome research: a proposal), where the target is typically a subunit of the rRNA gene (gene that codes for the ribosomal RNA): for instance, the V3-V4 region of the 16S subunit of this gene.
The amplification of the target sequence is usually obtained through a PCR (polymerase chain reaction) approach; sequencing can subsequently be performed with a next-generation or 3rd generation approach (you can have a look at Mestan et al. 2011, Quail et al. 2012 and Xiao & Zhou 2020).
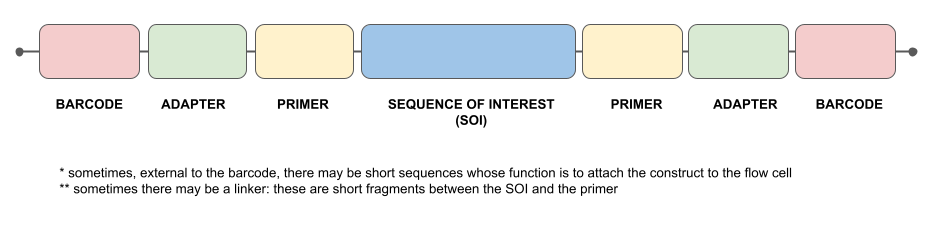
All right, but how do we construct our target sequence (a.k.a. sequence of interest, SOI) to be amplified and then sequenced? What about primers, barcodes, adapters? These names ring some bells, but what are they and where do they come into play in amplicon-seq? Here’s a super-simplified and super-quick sketch to clarify the matter.
- primer: the driving sequence that drives the amplification step: the (DNA-driven) polymerase enzyme recognises this sequence and copies (amplifies) the adjacent target sequence
- adapter: this is a sequence fragment that links the primer to the barcode
- barcode: this is a very short sequence (usually 6 or 8 bps) that tags the target sequence from each sample. The barcode allows to recognise from which sample each sequence is derived: after amplification and sequencing, we have millions of sequences which may be difficult to tell apart ( impossible if sequencing is in multiplexing, i.e. mixed sequences from multiple samples sequenced together).
The sketch below illustrates the relative positions of the barcode, adapter, primer and target sequence. Of course, everything is in duplicate (forward and reverse directions). [graphical work by Chiara Gini (Ph.D. student, University of Milan, 2022)]